Novel Unsupervised Feature Filtering of Biological Data
Supplementary Data
Roy
Varshavsky1*, Assaf Gottlieb2, Michal
Linial3 and David
Horn2
1School of Computer Science and Engineering, The
2School of Physics and Astronomy,
3Deptartment of Biological Chemistry,
1. The Viruses dataset of Fauquet, 1988
2. The MLL Leukemia dataset of Armstrong et al., 2002
3. The Leukemia dataset of Golub et al. 1999
4. GO Enrichment Tables
1. The Viruses dataset of Fauquet, 1988
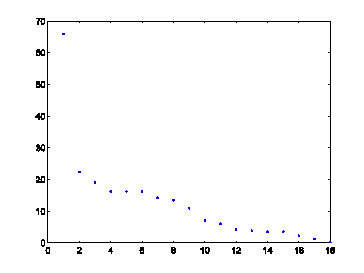
Figure 1: Variance of the features of the virus dataset
|
Spearman |
SR |
FS1 |
FS2 |
BE |
VS |
GS |
|
SR |
1 |
0.8824 |
0.63 |
0.8824 |
-0.0114 |
-0.4572 |
|
FS1 |
0.8824 |
1 |
0.4056 |
1 |
-0.2384 |
-0.676 |
|
FS2 |
0.63 |
0.4056 |
1 |
0.4056 |
0.4861 |
0.162 |
|
BE |
0.8824 |
1 |
0.4056 |
1 |
-0.2384 |
-0.676 |
|
VS |
-0.0114 |
-0.2384 |
0.4861 |
-0.2384 |
1 |
0.7647 |
|
GS |
-0.4572 |
-0.676 |
0.162 |
-0.676 |
0.7647 |
1 |
Table 1: Spearman correlation of the features ranking according to the various selection methods
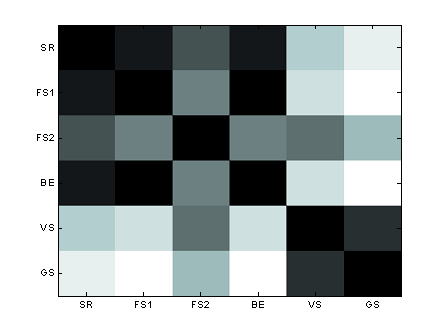
Figure 2: Spearman correlation of the features ranking according to the various selection methods
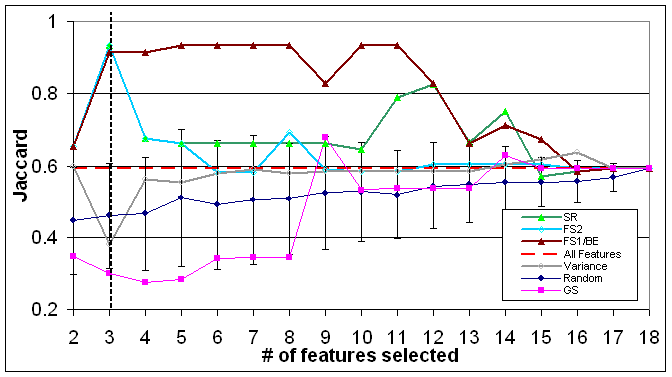
Figure 3: The quality of the various selection method of virus dataset (evaluation done by QC algorithm)
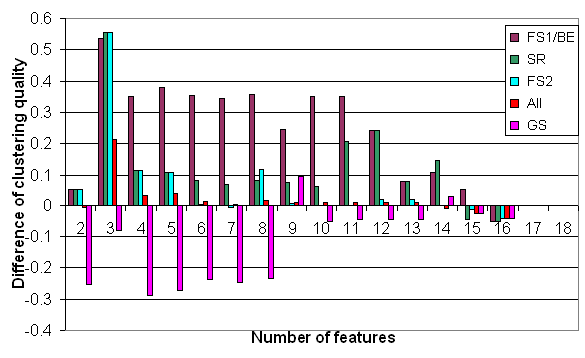
Figure 4:
Difference in clustering quality of the Virus
dataset, by selection according to the various selection methods. Displayed is
the difference from the variance selection (VS).
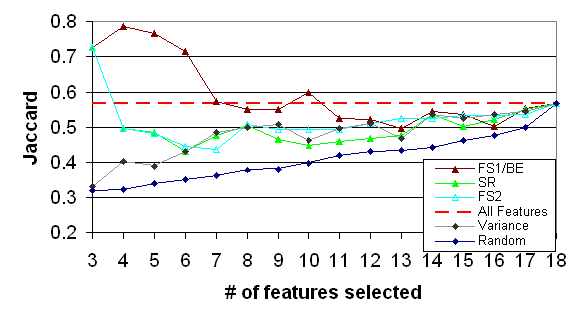
Figure 5: The quality of the various selection method of virus dataset (evaluation done by K-Means algorithm
2. The MLL Leukemia dataset of Armstrong et al., 2002
Figure 6: SR of the MLL dataset
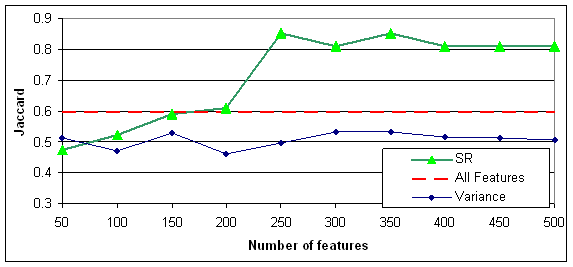
Figure 7: The quality of the various selection method of MLL dataset (evaluation done by QC algorithm
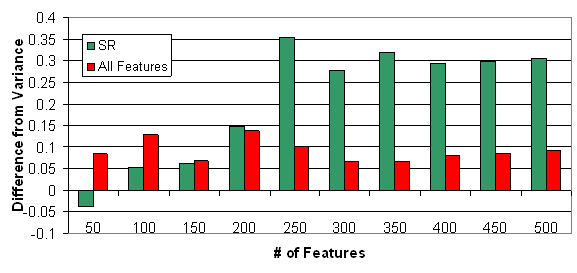
Figure 8: Difference in clustering quality of
the MLL dataset, by selection according to the various selection methods.
Displayed is the difference from the variance selection (VS).
3. The Leukemia dataset of Golub et al. 1999
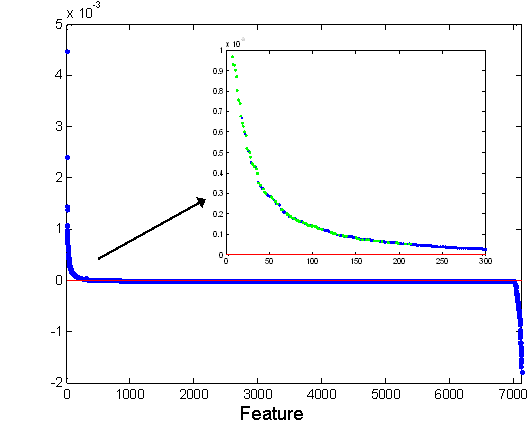
Figure 9: CE of the 7129 genes of the virus
dataset. The inset provides zoom into the highest-ranked 300 genes, with bright
dots signifying the top 100 features according to the FS1 method.
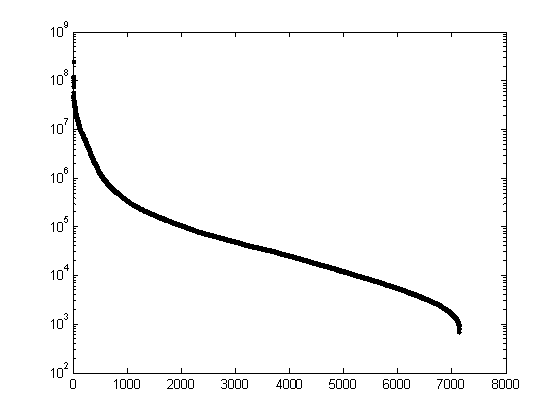
Figure 10: Log of the variance of the 7129 gene in the Leukemia dataset
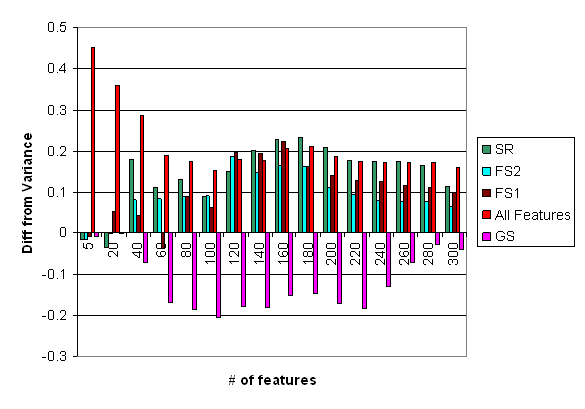
Figure 11: Difference in clustering quality of the Leukemia dataset, by selection according to the various selection methods. Displayed is the difference from the variance selection (VS).
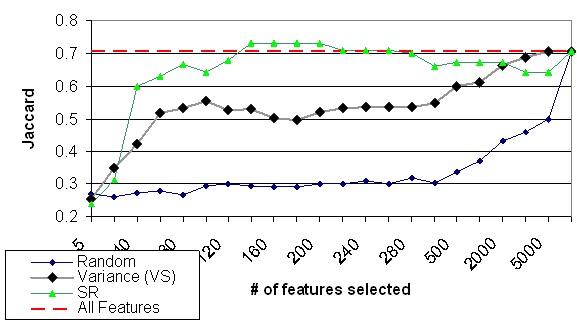
Figure 12: The quality of the various selection method of Golub dataset (evaluation done by QC algorithm
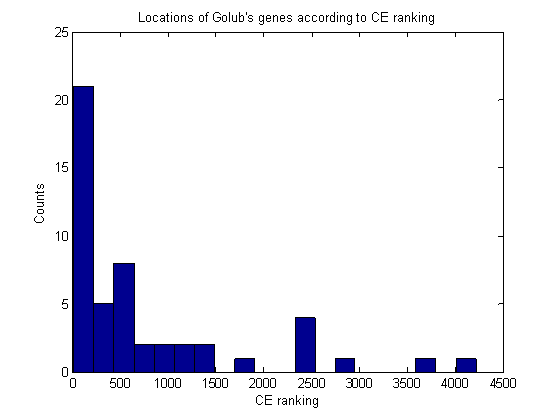
Figure 13: The location of Golub’s 50 genes on the CE ranking graph